Reopening Communities
Sun, April 26, 2020
Last month our world has been turned upside down. Many warm and welcoming communities used to large gatherings of people sharing hugs, meals and drinks, working on projects and playing games - implemented social distancing to help slow the spread of novel coronavirus. The Great Lockdown made impossible for communities to properly greet new neighbors, have wine parties and hike together as they had to adjust to life in a social distancing context.
The new plan to re-open the economy has been just released, but it will be a long road back to normal and it will have to pass through dystopian new normal.
Read more
Genes and Proteins of SARS-CoV-2
Mon, April 13, 2020
A pneumonia of unknown cause detected in Wuhan, China was
first reported to the WHO
Country Office in China on 31 December 2019.
 The outbreak was declared a Public Health Emergency of International Concern
on 30 January 2020 and declared a pandemic on March 11.
By that time the disease was known as COVID-19 and the name of the new virus was
SARS-CoV-2
The outbreak was declared a Public Health Emergency of International Concern
on 30 January 2020 and declared a pandemic on March 11.
By that time the disease was known as COVID-19 and the name of the new virus was
SARS-CoV-2
Researchers from Shanghai, Wuhan, Beijing and Sydney used metagenomic RNA sequencing to identify the previously unknown virus in a sample from "patient zero". This team deposited a draft genome sequence in the publicly available GenBank sequence repository on 10 January; the current version was deposited on 17 January.
A brief report published in New England Journal of Mediine on January 24 mentioned that the virus causing this disease had more than 85% identity with a bat SARS-like CoV (bat-SL-CoVZC45, MG772933.1).
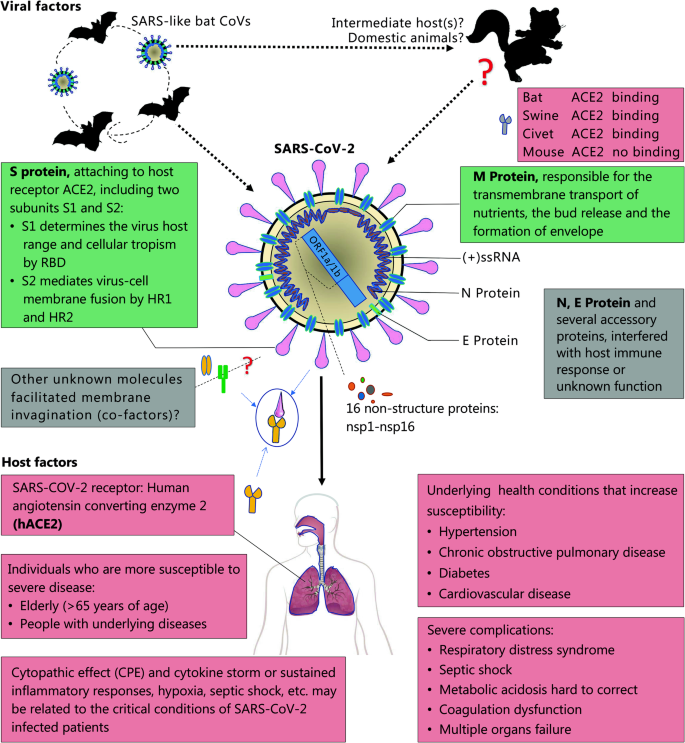
In early February, researchers published a paper describing the genome of SARS-Cov-2. Phylogenetic analysis of the complete viral genome revealed that it was most closely related (89.1% nucleotide similarity) to a group of SARS-like coronaviruses (genus Betacoronavirus, subgenus Sarbecovirus) found in bats in China. Later study reported 96.2% of whole-genome identity to BatCoV RaTG13 and suggested that pangolin species are a natural reservoir of SARS26 CoV-2-like CoVs.
To develop treatments, we need to learn more about viral proteins. A handful of genetic and structural analyses have identified a key feature of the virus — a protein on its surface — that might explain why it binds to and infects human cells so readily. The first structure of the receptor-binding domain from spike protein was solved and made available on January 1. Dozens more were published since. 3D maps were also created using cryo-electron microscopy. PDB data and related resources provided a starting point for structure-guided drug discovery and understanding of COVID-19.
In addition, Bioinformaticians identified all possible proteins that constitute Coronavirus and modeled their 3D structures. On March 5, DeepMind also made available their computational predictions of all structures associated with COVID-19. And Nvidia put out a call to PC gamers everywhere to donate their spare clock cycles toward advancing humanity's scientific knowledge of coronavirus. Next week, a new wave of projects "simulating potentially druggable protein targets from SARS-CoV-2 (the virus that causes COVID-19) and the related SARS-CoV virus (for which more structural data is available)" were made available on the service.
There are currently no FDA-approved treatments nor vaccines for coronavirus. But some approaches show promise. Researchers are working hard to produce precise, 3D molecular maps to guide the development of safe, effective ways of combating the pandemic. Scientists have already identified many new potential targets and compounds that can block the virus. We'll talk about it in the next blog.
REFERENCES
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML. A new coronavirus associated with human respiratory disease in China. Nature. 2020 Mar;579(7798):265-9.
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 doi: 10.1016/S0140-6736(20)30251-8.
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P. A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine. 2020 Jan 24.
Zhang T, Wu Q, Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Current Biology. 2020 Mar 19.
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Crystal structure of the 2019-nCoV spike receptor-binding domain bound with the ACE2 receptor. bioRxiv. 2020 Jan 1.
Srinivasan S, Cui H, Gao Z, Liu M, Lu S, Mkandawire W, Narykov O, Sun M, Korkin D. Structural Genomics of SARS-CoV-2 Indicates Evolutionary Conserved Functional Regions of Viral Proteins. Viruses. 2020 Apr;12(4):360. Sata available from
korkinlab.org/wuhan
Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AW, Bridgland A, Penedones H. Improved protein structure prediction using potentials from deep learning. Nature. 2020 Jan 15:1-5.
 The outbreak was declared a Public Health Emergency of International Concern
on 30 January 2020 and declared a pandemic on March 11.
By that time the disease was known as COVID-19 and the name of the new virus was
SARS-CoV-2
The outbreak was declared a Public Health Emergency of International Concern
on 30 January 2020 and declared a pandemic on March 11.
By that time the disease was known as COVID-19 and the name of the new virus was
SARS-CoV-2Researchers from Shanghai, Wuhan, Beijing and Sydney used metagenomic RNA sequencing to identify the previously unknown virus in a sample from "patient zero". This team deposited a draft genome sequence in the publicly available GenBank sequence repository on 10 January; the current version was deposited on 17 January.
A brief report published in New England Journal of Mediine on January 24 mentioned that the virus causing this disease had more than 85% identity with a bat SARS-like CoV (bat-SL-CoVZC45, MG772933.1).

In early February, researchers published a paper describing the genome of SARS-Cov-2. Phylogenetic analysis of the complete viral genome revealed that it was most closely related (89.1% nucleotide similarity) to a group of SARS-like coronaviruses (genus Betacoronavirus, subgenus Sarbecovirus) found in bats in China. Later study reported 96.2% of whole-genome identity to BatCoV RaTG13 and suggested that pangolin species are a natural reservoir of SARS26 CoV-2-like CoVs.
To develop treatments, we need to learn more about viral proteins. A handful of genetic and structural analyses have identified a key feature of the virus — a protein on its surface — that might explain why it binds to and infects human cells so readily. The first structure of the receptor-binding domain from spike protein was solved and made available on January 1. Dozens more were published since. 3D maps were also created using cryo-electron microscopy. PDB data and related resources provided a starting point for structure-guided drug discovery and understanding of COVID-19.
In addition, Bioinformaticians identified all possible proteins that constitute Coronavirus and modeled their 3D structures. On March 5, DeepMind also made available their computational predictions of all structures associated with COVID-19. And Nvidia put out a call to PC gamers everywhere to donate their spare clock cycles toward advancing humanity's scientific knowledge of coronavirus. Next week, a new wave of projects "simulating potentially druggable protein targets from SARS-CoV-2 (the virus that causes COVID-19) and the related SARS-CoV virus (for which more structural data is available)" were made available on the service.
There are currently no FDA-approved treatments nor vaccines for coronavirus. But some approaches show promise. Researchers are working hard to produce precise, 3D molecular maps to guide the development of safe, effective ways of combating the pandemic. Scientists have already identified many new potential targets and compounds that can block the virus. We'll talk about it in the next blog.
REFERENCES
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML. A new coronavirus associated with human respiratory disease in China. Nature. 2020 Mar;579(7798):265-9.
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 doi: 10.1016/S0140-6736(20)30251-8.
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P. A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine. 2020 Jan 24.
Zhang T, Wu Q, Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Current Biology. 2020 Mar 19.
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Crystal structure of the 2019-nCoV spike receptor-binding domain bound with the ACE2 receptor. bioRxiv. 2020 Jan 1.
Srinivasan S, Cui H, Gao Z, Liu M, Lu S, Mkandawire W, Narykov O, Sun M, Korkin D. Structural Genomics of SARS-CoV-2 Indicates Evolutionary Conserved Functional Regions of Viral Proteins. Viruses. 2020 Apr;12(4):360. Sata available from
korkinlab.org/wuhan
Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AW, Bridgland A, Penedones H. Improved protein structure prediction using potentials from deep learning. Nature. 2020 Jan 15:1-5.