Microbiome-directed medicine
Wednesday, September 30, 2020
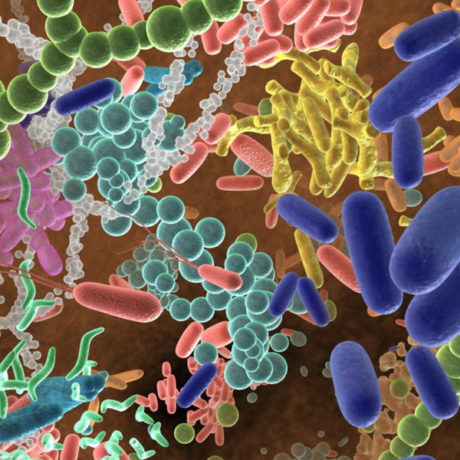
We live with hundred billions of roommates - microbes populating every nook and cranny in/on our body as well as our houses and cars. Some of them could be the best friends supporting our body as a happy household. They could be our defenders from hostile microorganisms, and suppliers of therapeutic compounds and tools for biotechnology. Yet, development of microbiome-directed medicine is stalled by the laborious nature of conventional cultivation methods and insufficient data for machine learning-driven approaches.
Petri dishes, flasks and culture tubes are no longer suitable for extensive characterization of specimens. Advances in single-cell and single-molecule technologies and microfluidics can facilitate the paradigm shift from single to multidisciplinary approaches intersecting biosciences, chemistry, and physics to accelerate the process. Arrays of miniaturized diffusion chambers, ‘isolation chips’ and ‘micro-Petri dishes’ being developed are promising tools of the future. Next challenge is to seamlessly integrate this with big data processing technologies.
Computational science and machine learning have a great potential to streamline microbiome-directed medicine. Newly developed bioinformatics tools are bringing opportunities in deciphering the microbiome data, from general-purpose sequence alignment and deep learning algorithms, to microbiome-specific approaches. New microbiome-related ‘omics’ datasets of ever increasing size and complexity are being produced. The application of AI for mining the microbiome requires the establishment of a global database repository with microbiome sequences and annotated host metadata. In addition, the use of co-measured features, such as metabolites and other ancillary information will significantly contribute to developing microbiome-based methods in precision medicine.
REFERENCES
- Ha CW, Devkota S. The new microbiology: Cultivating the future of microbiome-directed medicine. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2020 Sep 30.
- Su X, Jing G, Zhang Y, Wu S. Method development for cross-study microbiome data mining: Challenges and opportunities. Computational and Structural Biotechnology Journal. 2020 Aug 1.
- Zhou YH, Gallins P. A review and tutorial of machine learning methods for microbiome host trait prediction. Frontiers in Genetics. 2019;10:579.